Selected Projects
Soft Materials:
 Self-assembly of Dipolar Colloids: David Rutkowski and Ryan Maloney
Self-assembly of Dipolar Colloids: David Rutkowski and Ryan Maloney
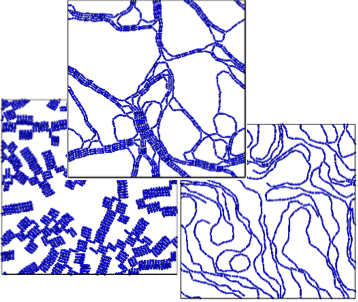
Discontinuous molecular dynamics and Monte Carlo simulations of 2-d systems of dipolar rods and of spheres with dipoles offset from the center line are conducted. Phase diagrams in the temperature – area fraction plane outlining the boundaries between fluid, string-fluid and percolated states are determined. The emphasis here is on network formation.
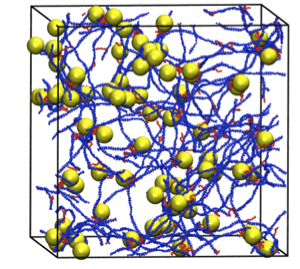 Simulations of Chitosan Assembly: Steven Benner
Simulations of Chitosan Assembly: Steven Benner
Chitosan is a versatile biopolymer with potential applications ranging from drug delivery and wound healing to flocculation of oil/water emulsions. Coarse-grained models of chitosan and hydrophobically modified chitosan have been developed that allow simulation via discontinuous molecular dynamics of the assembly of chitosan to form networks that encapsulate drug molecules.
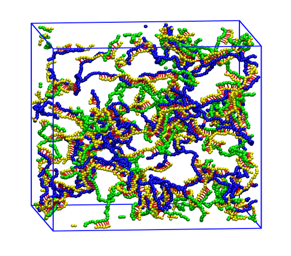 Aptamer-crosslinked Hydrogels: Kye Won Wang
Aptamer-crosslinked Hydrogels: Kye Won Wang
Aptamer-crosslinked hydrogels are being developed in the laboratory of our collaborator, T. Betancourt, for use as stimuli-responsive drug delivery systems. Coarse grained molecular dynamics simulations are being used to analyze their formation, drug loading capacity and degradation.
 Self-assembling Mixtures of Dipolar Rods and Spheres: Ryan Maloney
Self-assembling Mixtures of Dipolar Rods and Spheres: Ryan Maloney
Discontinuous molecular dynamics simulations of mixtures of dipolar rods and spheres reveal that they have much more complex behavior than is seen for either single component systems. Brownian dynamics simulations of mixtures of active and dipolar discs are also being conducted. These mixtures offer a potential path to creating new, temperature responsive, bifunctional materials.
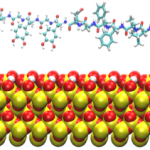 Underwater Adhesives: Amelia Chen and Qing Shao
Underwater Adhesives: Amelia Chen and Qing Shao
Studies on underwater organisms such as mussels and barnacles have shown that their natural glues involve two key components: L-3,4,-dihydroxyphenylalanine (L-DOPA) and amyloid nanostructures. Simulations are being used to investigate the behavior of L-DOPA and amyloid-forming peptides near surfaces, the goal being to design of synthetic, generic underwater adhesives.
 Protein Nanoparticle Interactions: Qing Shao
Protein Nanoparticle Interactions: Qing Shao
We investigated the allosteric effects of nanoparticles on proteins using a multiscale modeling approach and found that nanoparticles can change the properties of proteins even on regions distant from their binding site. This finding opens new ways to design nanoparticles for medical applications.
Protein Aggregation:
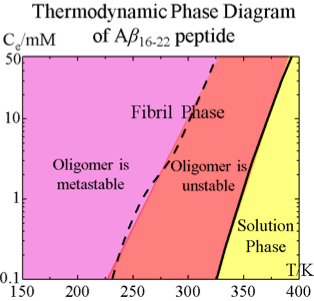 Aggregation of Aβ16-22 Peptides : Yiming Wang
Aggregation of Aβ16-22 Peptides : Yiming Wang
Aβ16-22 is a central hydrophobic fragment of Aβ protein. It’s aggregation kinetics is investigated by applying discontinuous molecular dynamics with the PRIME20 protein model. The thermodynamics of Aβ16-22 is revealed by constructing an equilibrium phase diagram in the temperature – concentration plane.

Inhibition of Aβ17-36 Aggregation : Yiming Wang
Some naturally occurring polyphenols (vanillin, resveratrol, curcumin and EGCG) can inhibit Aβ fibril formation and reduce neuron cell toxicity in vitro. Coarse-grained simulations were used to investigate their inhibition mechanism, helping us to understand the structure-function relationship underlying well-known amyloid inhibitors and design new ones.

Co-assembly of CATCH Peptides: Qing Shao
Peptide co-assembly is emerging as a new process to prepare nanomaterials. We are collaborating with experimentalists Gregory Hudalla (U Florida) and Anant Paravastu ( Georgia Tech) to understand the fundamentals that control peptide co-assembly and design high-performance co-assembling peptide systems.
Protein Design:
 Designing Peptides to Recognize Heart Attack Biomarkers: Xingqing Xiao
Designing Peptides to Recognize Heart Attack Biomarkers: Xingqing Xiao
A search algorithm was developed to design peptides that bind to specific biomolecular targets including cardiac troponin I, a heart attack biomarker in collaboration with R. Naik and P. Mirau at the Air Force Research lab. One in-silico peptide has ~16× higher affinity than the phage-display discovered peptide and a much lower limit of detection. The algorithm is being used to design sensors for cortisol.

Peptide Search Algorithms to Block Recruitment of tRNA-Lys3 by HIV: Xingqing Xiao
Blocking the recruitment of human tRNALys3, a primer for reverse transcription, by HIV has the potential to interfere with the virus life cycle. Our peptide search algorithm was used to design 15-residue peptides which could bind selectively to the anticodon stem and loop on tRNALys3 . When synthesized and characterized experimentally in the lab of our collaborator Paul Agris (Albany University) one of our designed peptides bound to the tRNALys3 with 10-fold higher affinity and specificity than the best phage-display identified peptide.